

 based on reviews by Mugino Kubo and 1 anonymous reviewer
based on reviews by Mugino Kubo and 1 anonymous reviewer
Three-dimensional microwear analysis is a very potent method in capturing the diet and, thus, reconstructing trophic relationships. It is widely applied in archaeology, palaeontology, neontology and (palaeo)anthropology. The method had been developed for mammal teeth (Walker et al., 1978; Teaford, 1988; Calandra and Merceron, 2016), but it has proven to be applicable to sharks (McLennan and Purnell, 2021) and reptiles, including fossil taxa with rather mysterious trophic ecologies (e.g., Bestwick et al., 2020; Holwerda et al., 2023). Microwear analysis has brought about landmark discoveries extending beyond autecology and reaching into palaeoenvironmental reconstructions (e.g., Merceron et al., 2016), niche evolution (e.g., Thiery et al., 2021), and assessment of food availability and niche partitioning (Ősi et al., 2022). Furthermore, microwear analysis is a testable method, which can be investigated experimentally in extant animals in order to ground-truth dietary interpretations in extinct organisms.
The study by Thiery et al. (2024) addresses important limitations of 3D microwear analysis: 1) the unequal access to commercial software required to analyze surface data obtained using confocal profilometers; 2) lack of replicability resulting from the use of commercial software with graphical user interface only. The latter point results in that documenting precisely what has been analyzed and how is nearly impossible.
The use of algorithms such as scale-sensitive fractal analysis (Ungar et al., 2003; Scott et al., 2006) and surface texture analysis has greatly improved replicability of DMTA and nearly eliminated intra- and inter-observer errors. Substantial effort has been made to quantify and minimize systematic and random errors in microwear analyses, such as intraspecific variation, use of different equipment (Arman et al., 2016), use of casts (Mihlbachler et al., 2019) or non-dietary variables (Bestwick et al., 2021). But even the best designed study cannot be replicated if the analysis is carried out with a “black box” software that many researchers may not afford. The trident package for R Software (https://github.com/nialsiG/trident) presented by Thiery et al. (2024) allows users to calculate 24 variables used in DMTA, transform them, calculate their variation across a surface, and rank them according to a sophisticated workflow that takes into account their normality and heteroscedasticity. A graphical user interface (GUI) is included in the form of a ShinyApp, but the power of the package, in my opinion, lies in that all steps of the analyses can be saved as R code and shared together with a study. This is a fundamental contribution to replicability and validation of microwear analyses. As best practices in code quality and replication become better known and accessible to palaeobiologists (The Turing Way Community, 2022; Trisovic et al., 2022). The presentation of the trident package is associated with three case studies, each with associated instructions on reproducing the results. These instructions partly use the literate programming approach, so that each step of the analysis is discussed and the methods are presented, either as screen shots when the GUI is used, or code. This is an excellent contribution, which hopefully will be followed by future microwear studies.
References
Arman, S. D., Ungar, P. S., Brown, C. A., DeSantis, L. R. G., Schmidt, C., and Prideaux, G. J. (2016). Minimizing inter-microscope variability in dental microwear texture analysis. Surface Topography: Metrology and Properties, 4(2), 024007. https://doi.org/10.1088/2051-672X/4/2/024007
Bestwick, J., Unwin, D. M., Butler, R. J., and Purnell, M. A. (2020). Dietary diversity and evolution of the earliest flying vertebrates revealed by dental microwear texture analysis. Nature Communications, 11(1), 5293. https://doi.org/10.1038/s41467-020-19022-2
Bestwick, J., Unwin, D. M., Henderson, D. M., and Purnell, M. A. (2021). Dental microwear texture analysis along reptile tooth rows: Complex variation with non-dietary variables. Royal Society Open Science, 8(2), 201754. https://doi.org/10.1098/rsos.201754
Calandra, I., and Merceron, G. (2016). Dental microwear texture analysis in mammalian ecology. Mammal Review, 46(3), 215–228. https://doi.org/10.1111/mam.12063
Holwerda, F. M., Bestwick, J., Purnell, M. A., Jagt, J. W. M., and Schulp, A. S. (2023). Three-dimensional dental microwear in type-Maastrichtian mosasaur teeth (Reptilia, Squamata). Scientific Reports, 13(1), 18720. https://doi.org/10.1038/s41598-023-42369-7
McLennan, L. J., and Purnell, M. A. (2021). Dental microwear texture analysis as a tool for dietary discrimination in elasmobranchs. Scientific Reports, 11(1), 2444. https://doi.org/10.1038/s41598-021-81258-9
Merceron, G., Novello, A., and Scott, R. S. (2016). Paleoenvironments inferred from phytoliths and Dental Microwear Texture Analyses of meso-herbivores. Geobios, 49(1–2), 135–146. https://doi.org/10.1016/j.geobios.2016.01.004
Mihlbachler, M. C., Foy, M., and Beatty, B. L. (2019). Surface replication, fidelity and data loss in traditional dental microwear and dental microwear texture analysis. Scientific Reports, 9(1), 1595. https://doi.org/10.1038/s41598-018-37682-5
Ősi, A., Barrett, P. M., Evans, A. R., Nagy, A. L., Szenti, I., Kukovecz, Á., Magyar, J., Segesdi, M., Gere, K., and Jó, V. (2022). Multi-proxy dentition analyses reveal niche partitioning between sympatric herbivorous dinosaurs. Scientific Reports, 12(1), 20813. https://doi.org/10.1038/s41598-022-24816-z
Scott, R. S., Ungar, P. S., Bergstrom, T. S., Brown, C. A., Childs, B. E., Teaford, M. F., and Walker, A. (2006). Dental microwear texture analysis: Technical considerations. Journal of Human Evolution, 51(4), 339–349. https://doi.org/10.1016/j.jhevol.2006.04.006
Teaford, M. F. (1988). A review of dental microwear and diet in modern mammals. Scanning Microscopy, 2, 1149–1166.
The Turing Way Community. (2022). The Turing Way: A handbook for reproducible, ethical and collaborative research (Version 1.0.2). Zenodo. https://doi.org/10.5281/ZENODO.3233853
Thiery, G., Francisco, A., Louail, M., Berlioz, É., Blondel, C., Brunetière, N., Ramdarshan, A., Walker, A. E. C., and Merceron, G. (2024). Introducing “trident”: A graphical interface for discriminating groups using dental microwear texture analysis. HAL, hal-04222508, ver. 4 peer-reviewed by PCI Paleo. https://hal.science/hal-04222508v4
Thiery, G., Gibert, C., Guy, F., Lazzari, V., Geraads, D., Spassov, N., and Merceron, G. (2021). From leaves to seeds? The dietary shift in late Miocene colobine monkeys of southeastern Europe. Evolution, 75(8), 1983–1997. https://doi.org/10.1111/evo.14283
Trisovic, A., Lau, M. K., Pasquier, T., and Crosas, M. (2022). A large-scale study on research code quality and execution. Scientific Data, 9(1), 60. https://doi.org/10.1038/s41597-022-01143-6
Ungar, P. S., Brown, C. A., Bergstrom, T. S., and Walker, A. (2003). Quantification of dental microwear by tandem scanning confocal microscopy and scale‐sensitive fractal analyses. Scanning, 25(4), 185–193. https://doi.org/10.1002/sca.4950250405
Walker, A., Hoeck, H. N., and Perez, L. (1978). Microwear of mammalian teeth as an indicator of diet. Science, 201(4359), 908–910. https://doi.org/10.1126/science.684415
DOI or URL of the preprint: https://hal.science/hal-04222508
Version of the preprint: 2
Dear colleagues,
Here are our responses for this final suggestions that we actually accepted
1. Thank you for this comment. The R Core Team and some packages package were cited in the Trident manual, where all used dependencies are mentioned, but they were not cited in the ms. This was of course an issue, and all the non-base packages are now cited in the manuscript, along with R Core Team.
2. The zenodo repository is now referenced and cited in teh Data availability part.
3. This is a good remark: we agree that .pdf is not the adequate format for sharing code. We can produce the R code as suggested (or the .Rmd file), but some contextual elements (figures) would be missing, making it difficult to follow (some steps must be performed in the UI, so it is not possible to run it all from the .R file anyway). Alternatively, we propose to upload an html file formatted with Rmarkdown: this way, the code can be copy-pasted and the contextual elements will help the reader follow the same process.
 , posted 13 Jul 2024, validated 15 Jul 2024
, posted 13 Jul 2024, validated 15 Jul 2024Dear Authors,
I am happy to let you know that the first reviewer has approved your modifications to the manuscript and found it ready for publication. Before I can recommend the preprint, I would like to ask you for a few final edits of technical nature. Attached please find the text with a few suggestions from my side. It shouldn't take longer than 5 min to apply them.
1. trident uses the PCA function of FactoMineR but FactoMineR is not referenced anywhere in the article not in the README of the GitHub repository. Please cite according to the authors of the package:
Lê, S., Josse, J. & Husson, F. (2008). FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software. 25(1). pp. 1-18.
along with any other dependencies. Not even R Software is cited in the manuscript. You can use e.g. the citation() function in R to see how to cite the Software and respective packages.
2. Please add (and cite) a reference to the Zenodo repository for the code: Ghislain Thiery, Francisco, A., Louail, M., & Merceron, G. (2023). nialsiG/trident: Trident 1.3.8 (trident). Zenodo. https://doi.org/10.5281/zenodo.8402605
3. I would like to emphasize in my recommendation the importance of script-based protocols for reproducibility of microwear analyses, but even the code used for the case studies is not shared in executable form, only inside a PDF, which means it cannot be run without extracting and correcting the line breaks. Would it be possible to share the scripts embedded in "Code_PCI_trident.pdf" as .R files?
I hope these suggestions will be easy to address and I am looking forward to seeing your article published with PCI Paleo.
Kind regards,
Emilia Jarochowska
Download recommender's annotations
Dear authors,
After reading your responses and the new version of the manuscript, I want to thank you for all the work you have undertaken. Congratulations for this new software and R package. I am sure it will be useful for many researchers.
I just have one last recommendation: please provide specific links or path to get the raw data on indores. I could not find them, even being myself a French speaker.
Looking forward seeing your manuscript in press,
Best regard,
https://doi.org/10.24072/pci.paleo.100256.rev21DOI or URL of the preprint: https://hal.science/hal-04222508
Version of the preprint: 1
Dear colleagues,
Please find below the revised version of the manuscript, including answers to comments and suggestions made by the reviewers. We also provide a link to the raw data accessible in InDores, a solution proposed by the French CNRS. These links are temporary (during review process) and will be open and available for the community once the paper will be (hopefully) accepted. Be aware that these links seem to not work for some Firefox versions.
Gildas MERCERON on behalf of all co-authors
++++++++++++++++++++++++++++++++++++++++++++++++
4 links for the Pig Experiment Data, Monkeys data, Modern Ruminants dataset from the Bauges Regional Park, and the extinct antelope dataset
https://data.indores.fr:443/privateurl.xhtml?token=2a85b83e-2cf7-42aa-80ec-baee7c905784- https://data.indores.fr:443/privateurl.xhtml?token=41e643b6-f389-4d70-b0ee-9cecd9c70ae5- https://data.indores.fr:443/privateurl.xhtml?token=fdf408d1-9997-48e6-89f4-5ed46e55256a- https://data.indores.fr:443/privateurl.xhtml?token=d300097a-f0ba-4850-b096-561019fade2a
 , posted 16 Dec 2023, validated 18 Dec 2023
, posted 16 Dec 2023, validated 18 Dec 2023Dear authors,
thank you for your submission of "Introducing ‘trident’: a graphical interface for discriminating groups using dental microwear texture analysis" and for your patience in recruiting reviewers and my own handling.
The two reviewers were positive about the importance of the package and the case studies you presented. I take that the main points in both reviews are some details missing that are needed to replicate your study. I do not have a Windows computer so it took me some time to install a Windows emulator to try trident and I wasn't able to reproduce the analyses: they are namely described in the text, but there is no code provided. It is, therefore, not clear if the analyses were carried out using the package's CLI or the Shiny App. If the former is the case, then the code for the analyses should also be provided. Further questions concerning the replicability of case studies were listed by reviewer 2.
As mentioned by reviewer 2, the structure of the manuscript, the focus is on the case studies and their scientific design and interpretation, but it is not always clear how much of the analyses is possible or different thanks to trident's functionalities. Could it be made more clear what about each case study is specific to trident?
Given trident's high utility, it is oddly difficult to use it; for example, it is not available on CRAN. This is perhaps because it only works on Windows. But also the instruction is not clear, e.g. it contains the sentence "Alternatively, you can follow the instructions in the readme.txt file." but I did not find any readme.txt file. Especially given its absence on CRAN, a complete README file on the package's repository, including a citation file, installation instructions etc, would be useful. While this is not in the manuscript's file, the manuscript is accompanying the software, therefore I include these comments here.
In terms of data handling, the final version of the manuscript should include the research data in a public repository, in compliance with FAIR principles.
Please consider citing all trident's dependencies. As fas as I can see only the citation for ggplot2 is included in the text, because it is used in the analyses. But the text should include a citation of trident itself (when it has a citation file), and citations - where applicable - of its dependencies should be at least in the README file to give due credit to the creators of research software (including yourselves).
I hope that you will find the comments by the reviewers constructive and useful. I would be grateful if you could address them and, if you disagree, explain why. I will be looking forward to reading the revised version of the manuscript.
Kind regards,
Emilia Jarochowska
 , 22 Nov 2023
, 22 Nov 2023Review comments to PCIPaleo #256 "Introducing ‘trident’: a graphical interface for discriminating groups using dental microwear texture analysis"
Thank you very much for the opportunity to review this very interesting paper. This research is closely related to my interests and the research that I am currently working on.
In this manuscript, a program package called "trident" developed by the authors for statistical comparison of dental microwear is introduced and its usefulness is demonstrated using three examples. As a researcher deeply involved in dental microwear research, I would like to express my sincere respect to the authors for developing such a wonderful program. This is because recent 3D dental microwear texture analysis (DMTA) involves the calculation of parameters that characterize microwear properties from the surface, but this computation is highly dependent on paid software (MountainsMap, sold by the French company DigitalSurf), which is a potential barrier to entry in this field (because of its high functionality, MountainsMap is very expensive).
While the paper is clearly written and has no major problems, I believe it is important to further clarify the reliability and usefulness of this program in order for it to be widely used, and some suggestions for manuscript revision are provided below. Some of these require additional calculations, graphing, figures, etc., and I encourage the authors to include them in the revised manuscript. Other minor comments are noted in the PDF.
I hope to see the revision published in PCIPaleo.
1) Prerequisite for trident
Authors explained that trident can read the SUR file, but on p. 9 L. 183-184 the authors write "All surfaces were preprocessed following Mercecron et al, (2016 )". It is necessary to clearly state whether software other than trident is required to prepare the SUR files for analysis and what further processing was done, rather than just showing a citation. This is important to indicate to the reader whether the entire analysis can be completed using only trident, or whether the data can only be used after it has been acquired with profilometers and then pre-processed with software other than trident.
2) Comparison of calculated values with the standard analysis software MountainsMap
As mentioned earlier, MountainsMap is widely used as the standard analysis software for DMTA. Therefore, if the values calculated with trident are shown to be consistent with those shown in previous studies, users who have already used MountainsMap can use trident with confidence. Specifically, for basic parameters such as Sq, it would be good to show that the values calculated by MountainsMap and those calculated by trident match on some (e.g. N=10) surface data (without subsampling of the surface).
3) Advantages of trident
The superiority of trident's new method (subsampling of the surface and then obtaining the dispersion indices of the parameters, which are then used as new parameters) over the traditional univariate comparison of DMTA parameters, or PCA with multiple parameters without ranking the variables should be shown. For example, Fig. 4C shows a box plot of PC1 using the trident method, but next to it is a box plot using univariate parameter (e.g. Sq), the variable with the largest significant difference between groups using the conventional method, or the conventional PCs, showing that trident's PC1 better captures the trend of the feeding groups. Similarly in Fig. 5, a comparison with the conventional method would be more convincing.
4) Screenshot of trident in use
The Supplement includes a manual for trident, which describes in detail the interface and usage of trident, but it would be easier for readers to understand what the program is like if the main manuscript also includes screenshots of trident in use.
None of the above comments require a great deal of effort on the part of the authors. Again, it is hoped that this software will lead to more DMTA research cases. The search for parameters that better illustrate differences in food properties (including derived parameters obtained by subsampling surfaces) will also provide a better understanding of how differences in food properties produce microwear features, i.e., the etiology of microwear. These future prospects should be discussed as a perspective at the end of the Discussion.
Download the review https://doi.org/10.24072/pci.paleo.100256.rev11General notes. The manuscript presents a new R package, and associated shiny application, called “trident”. The aim of this package is to analyse dental microwears using various measurements to finally detect which measurements are the best to discriminate the dataset into categories based on the diet, taxonomy etc. To illustrate the different applications of this package, the authors analysed three datasets. They detailed the analytical protocol and the results for each case (A, B and C) as well as how the results highlight the power of discrimination of trident.
What makes the trident R package of high interest is that it gathers various dental microwear texture analyses (DMTA) in one place. It considerably eases the investigations and the shiny application helps the users that are not used with R language, providing .SUR files as inputs. Even if I have a very positive opinion on this manuscript, I still have multiple questions and comments that are detailed below and I wish the authors answer to.
Abstract. It synthesises well the aim of the study in overall, but there are few information that are too detailed and few other information that are missing in my opinion. I would advise to remove the sentence about the remove of polynomial surfaces (l. 25-26). I don’t think specifying the number of variables and parameters (l. 26-27) is useful but it would be good to detail the “five different methods” (l. 28; isn’t it four instead, as said at l. 207?). Finally, I would advise the authors to add a first sentence that reminds the reader about the interest of dental microwear in ecological investigations.
Introduction. This section is well written in my opinion. I don’t think Figure 1 is necessary in the main text. Could it be replaced by a reference? Or put in supplementary information? More details are required to better understand this figure. Indeed, there is no information related to the studied taxa and their ecology. The units of each variable (epLsar and asfc) should also be explained.
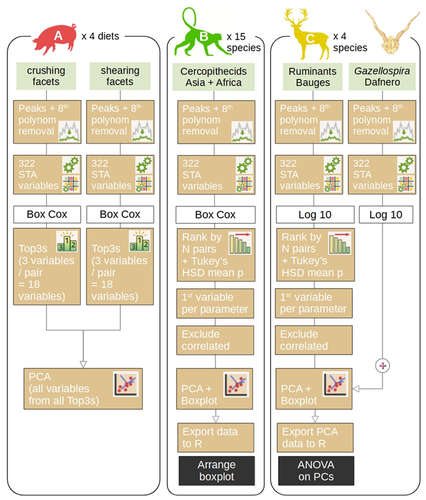
trident methodology. I think the flowchart in Figure 2 would be more useful if it depicted the whole methodology of trident package, especially when the variables are ranked depending their ability to discriminate categories. Each case A, B and C could then be applied on this general flowchart, using coloured arrows for example.
I am starting to get lost from lines 280 to 292. Why proposing all these protocols that have only subtle differences?
l. 280-283: I don’t understand the two sentences and so the protocol. What type of arrangement is it at line 280; what is this calculation of mean p-value at lines 281 and 282; what is the post-hoc tests at line 282; what p values are mentioned at line 283?
l. 284-285: what is “a given pair”? What is a “post-hoc p value”?
l.286-292: what data are used to perform Tukey’s HSD?
Case-specific analyses. To be honest, I have not fully understood the applied protocol for each case. A justification of the methodological choices is also missing: why choosing different protocols for these cases?
The authors keep the variables that are best to discriminate the categories in the different datasets. It is interesting to know which variables differentiate the most the categories, but what is finally the biological meaning of the variables? For me it is still a “jungle of parameters” if we cannot connect the variables to specific type of dental microwears and so to a specific diet. It would have been interested to compare the retained variables between the three cases to see the similarities/differences that could be imputed to the diet but also to the way the animals process the food. Perhaps the correlation circles in Figures 3-5 could be simplified in replacing the name of the variables by the type of microwear they refer to.
Case A.
l. 147 “… was fed 30% of barley seeds”: does it mean the diet was 70% base + 30% barley, or 100% base + 30% barley? I have the same question for cases B and C.
l. 307 “Variables that passed the multi check were classified”: what are the other variables and are they removed from the further classification? Wouldn’t it be better to favour a Kruskal-Wallis test to keep them in the analyses? Same question for cases B (l. 317) and C (l. 330-331).
l. 356 “the other groups are distinctly separated”: this can be tested using a MANOVA but it depends what the authors want to test, either a significant difference on the first PCA axes (even if it gathers only 60% of the variance), or a significant different based on the input variables that were used to build the PCA.
I cannot read Tables 3 and 4. What do the columns “p value ANOVA” and “Post-hoc p values” refer to? I don’t understand the ranking of the variables since they should be arranged based on their p-value: for example in Table 3, why Sk2 (p-value=0.04) is above Ssk (p-value=0.02) for the pair Co-CK?
l. 459: the separation between the groups has to be tested (see above).
Case B.
l. 323-324: why removing correlated parameters in this case (and in case C, l. 335-336) and not in case A? And why not integrating this step to the R script/trident protocol since it is used in 2/3 of the cases?
l. 385-394: since the diet of the taxa is one of the main focus of the results, I think it would be useful to add this information in Figure 4.
l. 483-484: if there is a continuum, then there is a significant difference between the two extremes that can be statistically tested. This continuum is observed only on PC1 which accounts for ~43% of the overall variance: the variables that contribute the most to this PC should be used as input to test this between-group difference.
l. 503: I don’t see any biomechanics in the discussion.
Case C. The choice of the different taxa is not justified in the main text: do they share phylogenetic relationships? Ecological similarities?
l. 337-338: “The remaining variables were used for a PCA. At this point, the surfaces of Gazellospira torticornis were added as supplementary individuals to the PCA”: so the retained variables for the extinct species are the same than the retained variables for the extant species?
l. 420-427: the authors mention significant differences between the extinct species and some of the extant taxa. These differences can be statistically tested with a MANOVA, based on the input variables that build the PCA.
Reproducibility. I was not able to use the shiny app as no .SUR file was available as supplementary material. I also tried to use trident package using the .TXT files given as supplementary material. The categories for each case is not easily available as I had to build data frames based on the information from Tables S1-S3. I found the function “trident.arrange” a bit cryptic since there is no detail about the available parameters for the argument “by”.
It would be great if the authors provided the script they used to analyse the data on R and/or the .SUR files so the reader could reproduce the results with the shiny app. The functions could be mentioned in the main text of the manuscript so the reader would know exactly what function to use for each methodological step.
Finally, as the retained variables are specific for each data set, how the analyses can be reproduced within a given taxonomic group? For example, if the data set of the domestic pigs is increased with new data (new specimens or even new diets), the retained variables might change. Then, it would be impossible to compare the new results with the old ones.
Additional remarks.
l. 114: “… and measure 16 variables”. I see only 15 variables in Table 2.
l. 230 and 231: I don’t understand this sentence. What are the 360 computed variables?
l.232-238: I don’t think Box.1 is useful since it is the only box in the whole manuscript. The text could be integrated in the main text.
l. 494-496: I don’t think this information is useful here.
https://doi.org/10.24072/pci.paleo.100256.rev12