
JAROCHOWSKA Emilia
- Department of Earth Sciences, Utrecht University, Utrecht, Netherlands
- Biomechanics & Functional morphology, Evolutionary biology, Evolutionary patterns and dynamics, Fieldwork, Fossilization, Fossil record, Geochemistry, Histology, Invertebrate paleontology, Macroecology, Macroevolution, Methods, Microfossils, Micropaleontology, Morphological evolution, Morphometrics, Paleobiodiversity, Paleobiology, Paleoecology, Paleoenvironments, Sclerochronology, Sedimentology, Systematics, Taphonomy, Taxonomy, Vertebrate paleontology
- recommender
Recommendations: 2
Reviews: 0
Recommendations: 2

Introducing ‘trident’: a graphical interface for discriminating groups using dental microwear texture analysis
A step towards improved replicability and accessibility of 3D microwear analyses
Recommended by Emilia Jarochowska based on reviews by Mugino Kubo and 1 anonymous reviewerThree-dimensional microwear analysis is a very potent method in capturing the diet and, thus, reconstructing trophic relationships. It is widely applied in archaeology, palaeontology, neontology and (palaeo)anthropology. The method had been developed for mammal teeth (Walker et al., 1978; Teaford, 1988; Calandra and Merceron, 2016), but it has proven to be applicable to sharks (McLennan and Purnell, 2021) and reptiles, including fossil taxa with rather mysterious trophic ecologies (e.g., Bestwick et al., 2020; Holwerda et al., 2023). Microwear analysis has brought about landmark discoveries extending beyond autecology and reaching into palaeoenvironmental reconstructions (e.g., Merceron et al., 2016), niche evolution (e.g., Thiery et al., 2021), and assessment of food availability and niche partitioning (Ősi et al., 2022). Furthermore, microwear analysis is a testable method, which can be investigated experimentally in extant animals in order to ground-truth dietary interpretations in extinct organisms.
The study by Thiery et al. (2024) addresses important limitations of 3D microwear analysis: 1) the unequal access to commercial software required to analyze surface data obtained using confocal profilometers; 2) lack of replicability resulting from the use of commercial software with graphical user interface only. The latter point results in that documenting precisely what has been analyzed and how is nearly impossible.
The use of algorithms such as scale-sensitive fractal analysis (Ungar et al., 2003; Scott et al., 2006) and surface texture analysis has greatly improved replicability of DMTA and nearly eliminated intra- and inter-observer errors. Substantial effort has been made to quantify and minimize systematic and random errors in microwear analyses, such as intraspecific variation, use of different equipment (Arman et al., 2016), use of casts (Mihlbachler et al., 2019) or non-dietary variables (Bestwick et al., 2021). But even the best designed study cannot be replicated if the analysis is carried out with a “black box” software that many researchers may not afford. The trident package for R Software (https://github.com/nialsiG/trident) presented by Thiery et al. (2024) allows users to calculate 24 variables used in DMTA, transform them, calculate their variation across a surface, and rank them according to a sophisticated workflow that takes into account their normality and heteroscedasticity. A graphical user interface (GUI) is included in the form of a ShinyApp, but the power of the package, in my opinion, lies in that all steps of the analyses can be saved as R code and shared together with a study. This is a fundamental contribution to replicability and validation of microwear analyses. As best practices in code quality and replication become better known and accessible to palaeobiologists (The Turing Way Community, 2022; Trisovic et al., 2022). The presentation of the trident package is associated with three case studies, each with associated instructions on reproducing the results. These instructions partly use the literate programming approach, so that each step of the analysis is discussed and the methods are presented, either as screen shots when the GUI is used, or code. This is an excellent contribution, which hopefully will be followed by future microwear studies.
References
Arman, S. D., Ungar, P. S., Brown, C. A., DeSantis, L. R. G., Schmidt, C., and Prideaux, G. J. (2016). Minimizing inter-microscope variability in dental microwear texture analysis. Surface Topography: Metrology and Properties, 4(2), 024007. https://doi.org/10.1088/2051-672X/4/2/024007
Bestwick, J., Unwin, D. M., Butler, R. J., and Purnell, M. A. (2020). Dietary diversity and evolution of the earliest flying vertebrates revealed by dental microwear texture analysis. Nature Communications, 11(1), 5293. https://doi.org/10.1038/s41467-020-19022-2
Bestwick, J., Unwin, D. M., Henderson, D. M., and Purnell, M. A. (2021). Dental microwear texture analysis along reptile tooth rows: Complex variation with non-dietary variables. Royal Society Open Science, 8(2), 201754. https://doi.org/10.1098/rsos.201754
Calandra, I., and Merceron, G. (2016). Dental microwear texture analysis in mammalian ecology. Mammal Review, 46(3), 215–228. https://doi.org/10.1111/mam.12063
Holwerda, F. M., Bestwick, J., Purnell, M. A., Jagt, J. W. M., and Schulp, A. S. (2023). Three-dimensional dental microwear in type-Maastrichtian mosasaur teeth (Reptilia, Squamata). Scientific Reports, 13(1), 18720. https://doi.org/10.1038/s41598-023-42369-7
McLennan, L. J., and Purnell, M. A. (2021). Dental microwear texture analysis as a tool for dietary discrimination in elasmobranchs. Scientific Reports, 11(1), 2444. https://doi.org/10.1038/s41598-021-81258-9
Merceron, G., Novello, A., and Scott, R. S. (2016). Paleoenvironments inferred from phytoliths and Dental Microwear Texture Analyses of meso-herbivores. Geobios, 49(1–2), 135–146. https://doi.org/10.1016/j.geobios.2016.01.004
Mihlbachler, M. C., Foy, M., and Beatty, B. L. (2019). Surface replication, fidelity and data loss in traditional dental microwear and dental microwear texture analysis. Scientific Reports, 9(1), 1595. https://doi.org/10.1038/s41598-018-37682-5
Ősi, A., Barrett, P. M., Evans, A. R., Nagy, A. L., Szenti, I., Kukovecz, Á., Magyar, J., Segesdi, M., Gere, K., and Jó, V. (2022). Multi-proxy dentition analyses reveal niche partitioning between sympatric herbivorous dinosaurs. Scientific Reports, 12(1), 20813. https://doi.org/10.1038/s41598-022-24816-z
Scott, R. S., Ungar, P. S., Bergstrom, T. S., Brown, C. A., Childs, B. E., Teaford, M. F., and Walker, A. (2006). Dental microwear texture analysis: Technical considerations. Journal of Human Evolution, 51(4), 339–349. https://doi.org/10.1016/j.jhevol.2006.04.006
Teaford, M. F. (1988). A review of dental microwear and diet in modern mammals. Scanning Microscopy, 2, 1149–1166.
The Turing Way Community. (2022). The Turing Way: A handbook for reproducible, ethical and collaborative research (Version 1.0.2). Zenodo. https://doi.org/10.5281/ZENODO.3233853
Thiery, G., Francisco, A., Louail, M., Berlioz, É., Blondel, C., Brunetière, N., Ramdarshan, A., Walker, A. E. C., and Merceron, G. (2024). Introducing “trident”: A graphical interface for discriminating groups using dental microwear texture analysis. HAL, hal-04222508, ver. 4 peer-reviewed by PCI Paleo. https://hal.science/hal-04222508v4
Thiery, G., Gibert, C., Guy, F., Lazzari, V., Geraads, D., Spassov, N., and Merceron, G. (2021). From leaves to seeds? The dietary shift in late Miocene colobine monkeys of southeastern Europe. Evolution, 75(8), 1983–1997. https://doi.org/10.1111/evo.14283
Trisovic, A., Lau, M. K., Pasquier, T., and Crosas, M. (2022). A large-scale study on research code quality and execution. Scientific Data, 9(1), 60. https://doi.org/10.1038/s41597-022-01143-6
Ungar, P. S., Brown, C. A., Bergstrom, T. S., and Walker, A. (2003). Quantification of dental microwear by tandem scanning confocal microscopy and scale‐sensitive fractal analyses. Scanning, 25(4), 185–193. https://doi.org/10.1002/sca.4950250405
Walker, A., Hoeck, H. N., and Perez, L. (1978). Microwear of mammalian teeth as an indicator of diet. Science, 201(4359), 908–910. https://doi.org/10.1126/science.684415
Morphometric changes in two Late Cretaceous calcareous nannofossil lineages support diversification fueled by long-term climate change
Insights into mechanisms of coccolithophore speciation: How useful is cell size in delineating species?
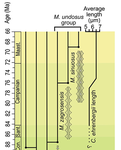
Recommended by Emilia Jarochowska based on reviews by Andrej Spiridonov and 1 anonymous reviewerCalcareous plankton gives us perhaps the most complete record of microevolutionary changes in the fossil record (e.g. Tong et al., 2018; Weinkauf et al., 2019), but this opportunity is not exploited enough, as it requires meticulous work in documenting assemblage-level variation through time. Especially in organisms such as coccolithophores, understanding the meaning of secular trends in morphology warrants an understanding of the functional biology and ecology of these organisms. Razmjooei and Thibault (2022) achieve this in their painstaking analysis of two coccolithophore lineages, Cribrosphaerella ehrenbergii and Microrhabdulus, in the Late Cretaceous of Iran. They propose two episodes of morphological change. The first one, starting around 76 Ma in the late Campanian, is marked by a sudden shift towards larger sizes of C. ehrenbergii and the appearance of a new species M. zagrosensis from M. undulatus. The second episode around 69 Ma (Maastrichtian) is inferred from a gradual size increase and morphological changes leading to probably anagenetic speciation of M. sinuosus n.sp.
The study remarkably analyzed the entire distributions of coccolith length and rod width, rather than focusing on summary statistics (De Baets et al., in press). This is important, because the range of variation determines the taxon’s evolvability with respect to the considered trait (Love et al., 2022). As the authors discuss, cell size in photosymbiotic unicellular organisms is not subject to the same constraints that will be familiar to researchers working e.g. on mammals (Niklas, 1994; Payne et al., 2009; Smith et al., 2016). Furthermore, temporal changes in size alone cannot be interpreted as evolutionary without knowledge of phenotypic plasticity and environmental clines present in the basin (Aloisi, 2015). The more important is that this study cross-tests size changes with other morphological parameters to examine whether their covariation supports inferred speciation events. The article addresses as well the effects of varying sedimentation rates (Hohmann, 2021) by, somewhat implicitly, correcting for the stratophenetic trend using an age-depth model and accounting for a hiatus. Such multifaceted approach as applied in this work is fundamental to unlock the dynamics of speciation offered by the microfossil record.
The study highlights also the link between shifts in size and diversity. Klug et al. (2015) have previously demonstrated that these two variables are related, as higher diversity is more likely to lead to extreme values of morphological traits, but this study suggests that the relationship is more intertwined: environmentally-driven rise in morphological variability (and thus in size) can lead to diversification. It is a fantastic illustration of the complexity of morphological evolution that, if it can be evaluated in terms of mechanisms, provides an insight into the dynamics of speciation.
References
Aloisi, G. (2015). Covariation of metabolic rates and cell size in coccolithophores. Biogeosciences, 12(15), 4665–4692. doi: 10.5194/bg-12-4665-2015
De Baets, K., Jarochowska, E., Buchwald, S. Z., Klug, C., and Korn, D. (In Press). Lithology controls ammonoid size distribution. Palaios.
Hohmann, N. (2021). Incorporating information on varying sedimentation rates into palaeontological analyses. PALAIOS, 36(2), 53–67. doi: 10.2110/palo.2020.038
Klug, C., De Baets, K., Kröger, B., Bell, M. A., Korn, D., and Payne, J. L. (2015). Normal giants? Temporal and latitudinal shifts of Palaeozoic marine invertebrate gigantism and global change. Lethaia, 48(2), 267–288. doi: 10.1111/let.12104
Love, A. C., Grabowski, M., Houle, D., Liow, L. H., Porto, A., Tsuboi, M., Voje, K.L., and Hunt, G. (2022). Evolvability in the fossil record. Paleobiology, 48(2), 186–209. doi: 10.1017/pab.2021.36
Niklas, K. J. (1994). Plant allometry: The scaling of form and process. Chicago: University of Chicago Press.
Payne, J. L., Boyer, A. G., Brown, J. H., Finnegan, S., Kowalewski, M., Krause, R. A., Lyons, S.K., McClain, C.R., McShea, D.W., Novack-Gottshall, P.M., Smith, F.A., Stempien, J.A., and Wang, S. C. (2009). Two-phase increase in the maximum size of life over 3.5 billion years reflects biological innovation and environmental opportunity. Proceedings of the National Academy of Sciences, 106(1), 24–27. doi: 10.1073/pnas.0806314106
Razmjooei, M. J., and Thibault, N. (2022). Morphometric changes in two Late Cretaceous calcareous nannofossil lineages support diversification fueled by long-term climate change. PaleorXiv, nfyc9, ver. 4, peer-reviewed by PCI Paleo. doi: 10.31233/osf.io/nfyc9
Smith, F. A., Payne, J. L., Heim, N. A., Balk, M. A., Finnegan, S., Kowalewski, M., Lyons, S.K., McClain, C.R., McShea, D.W., Novack-Gottshall, P.M., Anich, P.S., and Wang, S. C. (2016). Body size evolution across the Geozoic. Annual Review of Earth and Planetary Sciences, 44(1), 523–553. doi: 10.1146/annurev-earth-060115-012147
Tong, S., Gao, K., and Hutchins, D. A. (2018). Adaptive evolution in the coccolithophore Gephyrocapsa oceanica following 1,000 generations of selection under elevated CO2. Global Change Biology, 24(7), 3055–3064. doi: 10.1111/gcb.14065
Weinkauf, M. F. G., Bonitz, F. G. W., Martini, R., and Kučera, M. (2019). An extinction event in planktonic Foraminifera preceded by stabilizing selection. PLOS ONE, 14(10), e0223490. doi: 10.1371/journal.pone.0223490