
HLUSKO Leslea
- Paleobiology, Spanish National Center for Research on Human Evolution (CENIEH), Burgos, Spain
- Comparative anatomy, Evo-Devo, Evolutionary biology, Evolutionary patterns and dynamics, Evolutionary theory, Fieldwork, Fossil calibration, Fossilization, Fossil record, Histology, Macroecology, Macroevolution, Methods, Morphological evolution, Morphometrics, Paleoanthropology, Paleobiodiversity, Paleobiogeography, Paleobiology, Paleoecology, Paleoenvironments, Paleogenetics & Ancient DNA, Paleogeography, Phylogenetics, Systematics, Taphonomy, Taxonomy, Vertebrate paleontology
- recommender
Recommendation: 1
Reviews: 0
Recommendation: 1
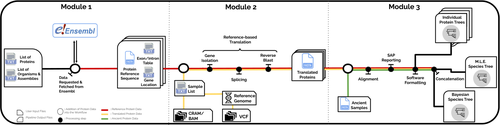
PaleoProPhyler: a reproducible pipeline for phylogenetic inference using ancient proteins
An open-source pipeline to reconstruct phylogenies with paleoproteomic data
Recommended by Leslea Hlusko based on reviews by Katerina Douka and 2 anonymous reviewersOne of the most recent technological advances in paleontology enables the characterization of ancient proteins, a new discipline known as palaeoproteomics (Ostrom et al., 2000; Warinner et al., 2022). Palaeoproteomics has superficial similarities with ancient DNA, as both work with ancient molecules, however the former focuses on peptides and the latter on nucleotides. While the study of ancient DNA is more established (e.g., Shapiro et al., 2019), palaeoproteomics is experiencing a rapid diversification of application, from deep time paleontology (e.g., Schroeter et al., 2022) to taxonomic identification of bone fragments (e.g., Douka et al., 2019), and determining genetic sex of ancient individuals (e.g., Lugli et al., 2022). However, as Patramanis et al. (2023) note in this manuscript, tools for analyzing protein sequence data are still in the informal stage, making the application of this methodology a challenge for many new-comers to the discipline, especially those with little bioinformatics expertise.
In the spirit of democratizing the field of palaeoproteomics, Patramanis et al. (2023) developed an open-source pipeline, PaleoProPhyler released under a CC-BY license (https://github.com/johnpatramanis/Proteomic_Pipeline). Here, Patramanis et al. (2023) introduce their workflow designed to facilitate the phylogenetic analysis of ancient proteins. This pipeline is built on the methods from earlier studies probing the phylogenetic relationships of an extinct genus of rhinoceros Stephanorhinus (Cappellini et al., 2019), the large extinct ape Gigantopithecus (Welker et al., 2019), and Homo antecessor (Welker et al., 2020). PaleoProPhyler has three interacting modules that initialize, construct, and analyze an input dataset. The authors provide a demonstration of application, presenting a molecular hominid phyloproteomic tree.
In order to run some of the analyses within the pipeline, the authors also generated the Hominid Palaeoproteomic Reference Dataset which includes 10,058 protein sequences per individual translated from publicly available whole genomes of extant hominids (orangutans, gorillas, chimpanzees, and humans) as well as some ancient genomes of Neanderthals and Denisovans. This valuable research resource is also publicly available, on Zenodo (Patramanis et al., 2022).
Three reviewers reported positively about the development of this program, noting its importance in advancing the application of palaeoproteomics more broadly in paleontology.
References
Cappellini, E., Welker, F., Pandolfi, L., Ramos-Madrigal, J., Samodova, D., Rüther, P. L., Fotakis, A. K., Lyon, D., Moreno-Mayar, J. V., Bukhsianidze, M., Rakownikow Jersie-Christensen, R., Mackie, M., Ginolhac, A., Ferring, R., Tappen, M., Palkopoulou, E., Dickinson, M. R., Stafford, T. W., Chan, Y. L., … Willerslev, E. (2019). Early Pleistocene enamel proteome from Dmanisi resolves Stephanorhinus phylogeny. Nature, 574(7776), 103–107. https://doi.org/10.1038/s41586-019-1555-y
Douka, K., Brown, S., Higham, T., Pääbo, S., Derevianko, A., and Shunkov, M. (2019). FINDER project: Collagen fingerprinting (ZooMS) for the identification of new human fossils. Antiquity, 93(367), e1. https://doi.org/10.15184/aqy.2019.3
Lugli, F., Nava, A., Sorrentino, R., Vazzana, A., Bortolini, E., Oxilia, G., Silvestrini, S., Nannini, N., Bondioli, L., Fewlass, H., Talamo, S., Bard, E., Mancini, L., Müller, W., Romandini, M., and Benazzi, S. (2022). Tracing the mobility of a Late Epigravettian (~ 13 ka) male infant from Grotte di Pradis (Northeastern Italian Prealps) at high-temporal resolution. Scientific Reports, 12(1), 8104. https://doi.org/10.1038/s41598-022-12193-6
Ostrom, P. H., Schall, M., Gandhi, H., Shen, T.-L., Hauschka, P. V., Strahler, J. R., and Gage, D. A. (2000). New strategies for characterizing ancient proteins using matrix-assisted laser desorption ionization mass spectrometry. Geochimica et Cosmochimica Acta, 64(6), 1043–1050. https://doi.org/10.1016/S0016-7037(99)00381-6
Patramanis, I., Ramos-Madrigal, J., Cappellini, E., and Racimo, F. (2022). Hominid Palaeoproteomic Reference Dataset (1.0.1) [dataset]. Zenodo. https://doi.org/10.5281/ZENODO.7333226
Patramanis, I., Ramos-Madrigal, J., Cappellini, E., and Racimo, F. (2023). PaleoProPhyler: A reproducible pipeline for phylogenetic inference using ancient proteins. BioRxiv, 519721, ver. 3 peer-reviewed by PCI Paleo. https://doi.org/10.1101/2022.12.12.519721
Schroeter, E. R., Cleland, T. P., and Schweitzer, M. H. (2022). Deep Time Paleoproteomics: Looking Forward. Journal of Proteome Research, 21(1), 9–19. https://doi.org/10.1021/acs.jproteome.1c00755
Shapiro, B., Barlow, A., Heintzman, P. D., Hofreiter, M., Paijmans, J. L. A., and Soares, A. E. R. (Eds.). (2019). Ancient DNA: Methods and Protocols (2nd ed., Vol. 1963). Humana, New York. https://doi.org/10.1007/978-1-4939-9176-1
Warinner, C., Korzow Richter, K., and Collins, M. J. (2022). Paleoproteomics. Chemical Reviews, 122(16), 13401–13446. https://doi.org/10.1021/acs.chemrev.1c00703
Welker, F., Ramos-Madrigal, J., Gutenbrunner, P., Mackie, M., Tiwary, S., Rakownikow Jersie-Christensen, R., Chiva, C., Dickinson, M. R., Kuhlwilm, M., De Manuel, M., Gelabert, P., Martinón-Torres, M., Margvelashvili, A., Arsuaga, J. L., Carbonell, E., Marques-Bonet, T., Penkman, K., Sabidó, E., Cox, J., … Cappellini, E. (2020). The dental proteome of Homo antecessor. Nature, 580(7802), 235–238. https://doi.org/10.1038/s41586-020-2153-8
Welker, F., Ramos-Madrigal, J., Kuhlwilm, M., Liao, W., Gutenbrunner, P., De Manuel, M., Samodova, D., Mackie, M., Allentoft, M. E., Bacon, A.-M., Collins, M. J., Cox, J., Lalueza-Fox, C., Olsen, J. V., Demeter, F., Wang, W., Marques-Bonet, T., and Cappellini, E. (2019). Enamel proteome shows that Gigantopithecus was an early diverging pongine. Nature, 576(7786), 262–265. https://doi.org/10.1038/s41586-019-1728-8