
DE BAETS Kenneth
- Faculty of Biology, Institute of Evolutionary Biology, University of Warsaw, Warsaw, Poland
- Biomechanics & Functional morphology, Biostratigraphy, Evo-Devo, Evolutionary biology, Evolutionary patterns and dynamics, Evolutionary theory, Fieldwork, Fossil calibration, Fossilization, Fossil record, Geochemistry, Histology, Index fossils, Invertebrate paleontology, Macroecology, Macroevolution, Methods, Morphological evolution, Morphometrics, Paleobiodiversity, Paleobiogeography, Paleobiology, Paleoecology, Paleoenvironments, Paleogeography, Paleopathology, Phylogenetics, Systematics, Taphonomy, Taxonomy
- recommender
Recommendation: 1
Reviews: 2
Recommendation: 1

Ammonoid taxonomy with supervised and unsupervised machine learning algorithms
Performance of machine-learning approaches in identifying ammonoid species based on conch properties
Recommended by Kenneth De Baets based on reviews by Jérémie Bardin and 1 anonymous reviewerThere are less and less experts on taxonomy of particular groups particularly among early career paleontologists and (paleo)biologists – this also includes ammonoid cephalopods. Techniques cannot replace this taxonomic expertise (Engel et al. 2021) but machine learning approaches can make taxonomy more efficient, reproducible as well as passing it over more sustainable. Initially ammonoid taxonomy was a black box with small differences sometimes sufficient to erect different species as well as really idiosyncratic groupings of superficially similar specimens (see De Baets et al. 2015 for a review). In the meantime, scientists have embraced more quantitative assessments of conch shape and morphology more generally (see Klug et al. 2015 for a more recent review). The approaches still rely on important but time-intensive collection work and seeing through daisy chains of more or less accessible papers and monographs without really knowing how these approaches perform (other than expert opinion). In addition, younger scientists are usually trained by more experienced scientists, but this practice is becoming more and more difficult which makes it difficult to resolve the taxonomic gap. This relates to the fact that less and less experienced researchers with this kind of expertise get employed as well as graduate students or postdocs choosing different research or job avenues after their initial training effectively leading to a leaky pipeline and taxonomic impediment.
Robust taxonomy and stratigraphy is the basis for all other studies we do as paleontologists/paleobiologists so Foxon (2021) represents the first step to use supervised and unsupervised machine-learning approaches and test their efficiency on ammonoid conch properties. This pilot study demonstrates that machine learning approaches can be reasonably accurate (60-70%) in identifying ammonoid species (Foxon, 2021) – at least similar to that in other mollusk taxa (e.g., Klinkenbuß et al. 2020) - and might also be interesting to assist in cases where more traditional methods are not feasible. Novel approaches might even allow to further approve the accuracy as has been demonstrated for other research objects like pollen (Romero et al. 2020). Further applying of machine learning approaches on larger datasets and additional morphological features (e.g., suture line) are now necessary in order to test and improve the robustness of these approaches for ammonoids as well as test their performance more broadly within paleontology.
References
De Baets K, Bert D, Hoffmann R, Monnet C, Yacobucci M, and Klug C (2015). Ammonoid intraspecific variability. In: Ammonoid Paleobiology: From anatomy to ecology. Ed. by Klug C, Korn D, De Baets K, Kruta I, and Mapes R. Vol. 43. Topics in Geobiology. Dordrecht: Springer, pp. 359–426.
Engel MS, Ceríaco LMP, Daniel GM, Dellapé PM, Löbl I, Marinov M, Reis RE, Young MT, Dubois A, Agarwal I, Lehmann A. P, Alvarado M, Alvarez N, Andreone F, Araujo-Vieira K, Ascher JS, Baêta D, Baldo D, Bandeira SA, Barden P, Barrasso DA, Bendifallah L, Bockmann FA, Böhme W, Borkent A, Brandão CRF, Busack SD, Bybee SM, Channing A, Chatzimanolis S, Christenhusz MJM, Crisci JV, D’elía G, Da Costa LM, Davis SR, De Lucena CAS, Deuve T, Fernandes Elizalde S, Faivovich J, Farooq H, Ferguson AW, Gippoliti S, Gonçalves FMP, Gonzalez VH, Greenbaum E, Hinojosa-Díaz IA, Ineich I, Jiang J, Kahono S, Kury AB, Lucinda PHF, Lynch JD, Malécot V, Marques MP, Marris JWM, Mckellar RC, Mendes LF, Nihei SS, Nishikawa K, Ohler A, Orrico VGD, Ota H, Paiva J, Parrinha D, Pauwels OSG, Pereyra MO, Pestana LB, Pinheiro PDP, Prendini L, Prokop J, Rasmussen C, Rödel MO, Rodrigues MT, Rodríguez SM, Salatnaya H, Sampaio Í, Sánchez-García A, Shebl MA, Santos BS, Solórzano-Kraemer MM, Sousa ACA, Stoev P, Teta P, Trape JF, Dos Santos CVD, Vasudevan K, Vink CJ, Vogel G, Wagner P, Wappler T, Ware JL, Wedmann S, and Zacharie CK (2021). The taxonomic impediment: a shortage of taxonomists, not the lack of technical approaches. Zoological Journal of the Linnean Society 193, 381–387. doi: 10. 1093/zoolinnean/zlab072
Foxon F (2021). Ammonoid taxonomy with supervised and unsupervised machine learning algorithms. PaleorXiv ewkx9, ver. 3, peer-reviewed by PCI Paleo. doi: 10.31233/osf.io/ewkx9
Klinkenbuß D, Metz O, Reichert J, Hauffe T, Neubauer TA, Wesselingh FP, and Wilke T (2020). Performance of 3D morphological methods in the machine learning assisted classification of closely related fossil bivalve species of the genus Dreissena. Malacologia 63, 95. doi: 10.4002/040.063.0109
Klug C, Korn D, Landman NH, Tanabe K, De Baets K, and Naglik C (2015). Ammonoid conchs. In: Ammonoid Paleobiology: From anatomy to ecology. Ed. by Klug C, Korn D, De Baets K, Kruta I, and Mapes RH. Vol. 43. Dordrecht: Springer, pp. 3–24.
Romero IC, Kong S, Fowlkes CC, Jaramillo C, Urban MA, Oboh-Ikuenobe F, D’Apolito C, and Punyasena SW (2020). Improving the taxonomy of fossil pollen using convolutional neural networks and superresolution microscopy. Proceedings of the National Academy of Sciences 117, 28496–28505. doi: 10.1073/pnas.2007324117
Reviews: 2
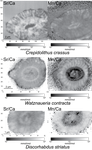
Evidence of high Sr/Ca in a Middle Jurassic murolith coccolith species
New results and challenges in Sr/Ca studies on Jurassic coccolithophorids
Recommended by Antonino Briguglio based on reviews by Kenneth De Baets and 1 anonymous reviewerThis interesting publication by Suchéras-Marx et al. (2020) highlights peculiar aspects of geochemistry in nannofossils, specifically coccolithophorids. One of the main application of geochemistry on fossil shells is to get hints on the physiology of such extinct taxa. Here, the authors try to get information on the calcification mechanism and processes in Jurassic coccoliths. Coccoliths build a test made of calcium carbonate and one of the most common geochemical proxies used for this fossil group is the Sr/Ca ratio. This isotopic ratio has good chances to be successfully used as a robust proxy for paleoenvironmental reconstruction, but, concerning Jurassic coccoliths things seem to be not straightforward.
The authors managed to compare the isotopic value of Sr/Ca measured on Jurassic coccoliths from different taxonomic groups: the murolith Crepidolithus crassus and the placoliths Watznaueria contracta and Discorhabdus striatus. The results they got clearly show that the Sr/Ca ratio cannot be used as a universal proxy because these species exhibit very different values despite coming from the same stratigraphic level and having undergone minimal diagenetic modification. Data seem to point to a Sr/Ca ratio up to 10 times higher in the murolith species than in the placolith taxa (Suchéras-Marx et al., 2020). One of the explanation given here takes advantage of modern coccolith data and hints to specific polysaccharides that would control the growth of the long R unit in the murolith species. As always, there is plenty of space for additional research, possibly on modern taxa, to sort out the scientific questions that arise from this work.
References
Suchéras-Marx, B., Giraud, F., Simionovici, A., Tucoulou, R., & Daniel, I. (2020). Evidence of high Sr/Ca in a Middle Jurassic murolith coccolith species. PaleorXiv, dcfuq, version 7, peer-reviewed by PCI Paleo. doi: 10.31233/osf.io/dcfuq

A simple generative model of trilobite segmentation and growth
Deep insights into trilobite development
Recommended by Christian Klug based on reviews by Kenneth De Baets and Lukas LaiblTrilobites are arthropods that became extinct at the greatest marine mass extinction over 250 Ma ago. Because of their often bizarre forms, their great diversity and disparity of shapes, they have attracted the interest of researchers and laypersons alike. Due to their calcified exoskeleton, their remains are quite abundant in many marine strata. One particularly interesting aspect, however, is the fossilization of various molting stages. This allows the reconstruction of both juvenile strategies (lecitotrophic versus planktotrophic) and the entire life history of at least some well-documented taxa (e.g., Hughes 2003, 2007; Laibl 2017). For example, life history of trilobites is, based on certain morphological changes, classically subdivided in the three phases protaspis (hatchling, one dorsal shield with few segments with no articulation between), meraspis (juvenile, two and more shields connected by articulations) and holaspis (when the terminal number of thoracic segments is reached). At most molting events, a new skeletal element is added (only in the holaspis, the number of thoracic segments does not change). Nevertheless, many trilobites are known mainly from late meraspid and holaspid stages, because the dorsal shields of the first ontogenetic stages are usually very small and thus often either dissolved or overlooked. An improved understanding of trilobite ontogeny could thus help filling in these gaps in fossil preservation and subsequently, to better understand evolutionary pathways. This is where this paper comes in. In a very clever approach, the New-York-based researcher Melanie Hopkins modeled the growth of these segmented animals (Hopkins 2020). Previous growth models of invertebrates focused on, e.g., mollusks, whose shells grow by accretion. Modelling arthropod ontogeny represented a challenge, which is now overcome by Hopkins' brilliant paper. Her generative growth model is based on empirical data of *Aulacopleura koninckii* (Barrande, 1846). Hong et al. (2014) and Hughes et al. (2017) documented the ontogeny of this 429 Ma old trilobite species in great detail. In the Silurian of the Barrandian region (Czech Republic), this species is locally very common and all growth stages are well known. I could imagine that the paper of Hughes et al. (2017) planted the seed into Melanie Hopkins’ mind to approach trilobite development in general in a quantitative way with a mathematical approach comparable to the mollusk-research by, e.g., David Raup (1961, 1966) and George McGhee (2015). Hopkins’ growth model requires “a minimum of nine parameters […] to model basic trilobite growth and segmentation, and three additional parameters […] to allow a transition to a new growth gradient for the trunk region during ontogeny” (Hopkins 2020: p. 21). It is now possible to play with parameters such as protaspid size, segment dimensions, segment numbers, etc., in order to estimate changes in body size or morphology. Furthermore, the model could be applied to similarly organized arthropod exoskeletons like many early Cambrian arthropods (e.g., marellomorphs) or even crustaceans (e.g., conchostracans or copepods). Of great interest could also be to assess influences of environmental changes on arthropod ontogeny. Also, her work will help to reconstruct unknown developmental information missing from trilobite species (and possibly other arthropods) and also to explore their morphospace. **References** Barrande, J. (1846). Notice préliminaire sur le système Silurien et les trilobites de Bohême. Leipzig: Hirschfield. Hong, P. S., Hughes, N. C., & Sheets, H. D. (2014). Size, shape, and systematics of the Silurian trilobite *Aulacopleura koninckii*. Journal of Paleontology, 88(6), 1120–1138. doi: [ 10.1666/13-142](https://dx.doi.org/ 10.1666/13-142) Hopkins, M. J. (2020). A simple generative model of trilobite segmentation and growth. PaleorXiv, version 3, peer-reviewed by PCI Paleo. doi: [ 10.31233/osf.io/zt642](https://dx.doi.org/ 10.31233/osf.io/zt642) Hughes, N. C. (2003). Trilobite tagmosis and body patterning from morphological and developmental perspectives. Integrative and Comparative Biology, 43(1), 185–206. doi: [ 10.1093/icb/43.1.185](https://dx.doi.org/ 10.1093/icb/43.1.185) Hughes, N. C. (2007). The evolution of trilobite body patterning. Annual Review of Earth and Planetary Sciences, 35(1), 401–434. doi: [ 10.1146/annurev.earth.35.031306.140258](https://dx.doi.org/ 10.1146/annurev.earth.35.031306.140258) Hughes, N. C., Hong, P. S., Hou, J., & Fusco, G. (2017). The development of the Silurian trilobite *Aulacopleura koninckii* reconstructed by applying inferred growth and segmentation dynamics: A case study in paleo-evo-devo. Frontiers in Ecology and Evolution, 5, 00037. doi: [ 10.3389/fevo.2017.00037](https://dx.doi.org/ 10.3389/fevo.2017.00037) Laibl, L. (2017). Patterns in Palaeontology: The development of trilobites. Palaeontology Online, 7(10), 1–9. McGhee, G. R. (2015). Limits in the evolution of biological form: a theoretical morphologic perspective. Interface Focus, 5(6), 20150034. doi: [ 10.1098/rsfs.2015.0034](https://dx.doi.org/ 10.1098/rsfs.2015.0034) Raup, D. M. (1961). The geometry of coiling in gastropods. Proceedings of the National Academy of Sciences, 47(4), 602–609. doi: [ 10.1073/pnas.47.4.602](https://dx.doi.org/ 10.1073/pnas.47.4.602) Raup, D. M. (1966). Geometric analysis of shell coiling: general problems. Journal of Paleontology, 40, 1178–1190.